Hybrid graph convolutional neural networks for landmark-based anatomical segmentation
Published:
Gaggion, N., Mansilla, L., Milone, D. H., & Ferrante, E. (2021, September). Hybrid graph convolutional neural networks for landmark-based anatomical segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 600-610). Springer, Cham. https://link.springer.com/chapter/10.1007/978-3-030-87193-2_57
Hybrid graph convolutional neural networks for landmark-based anatomical segmentation
Abstract
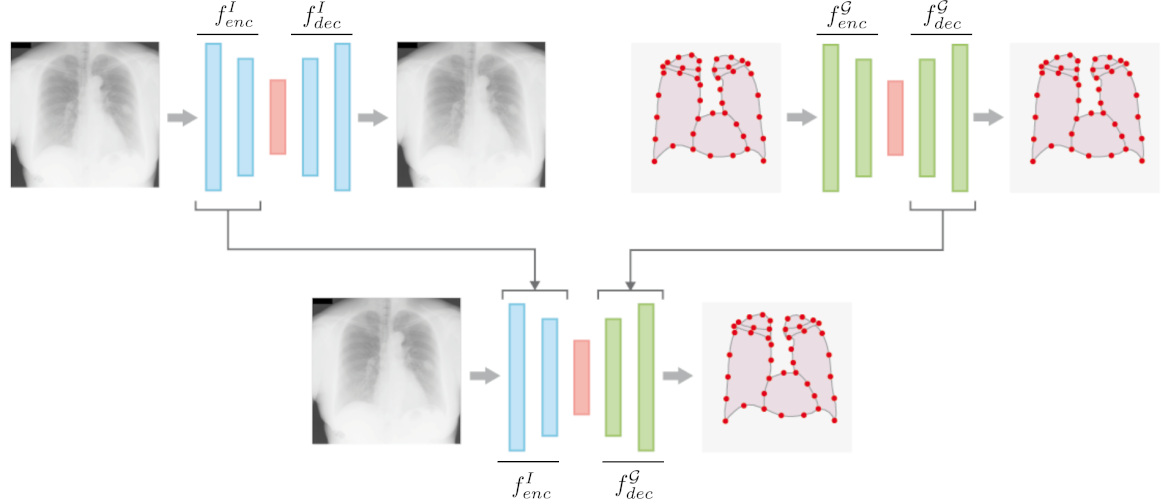
In this work we address the problem of landmark-based segmentation for anatomical structures. We propose HybridGNet, an encoder-decoder neural architecture which combines standard convolutions for image feature encoding, with graph convolutional neural networks to decode plausible representations of anatomical structures. We benchmark the proposed architecture considering other standard landmark and pixel-based models for anatomical segmentation in chest x-ray images, and found that HybridGNet is more robust to image occlusions. We also show that it can be used to construct landmark-based segmentations from pixel level annotations. Our experimental results suggest that Hybrid-Net produces accurate and anatomically plausible landmark-based segmentations, by naturally incorporating shape constraints within the decoding process via spectral convolutions.
Citation
@InProceedings{10.1007/978-3-030-87193-2_57,
author="Gaggion, Nicol{\'a}s
and Mansilla, Lucas
and Milone, Diego H.
and Ferrante, Enzo",
title="Hybrid Graph Convolutional Neural Networks for Landmark-Based Anatomical Segmentation",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2021",
year="2021",
publisher="Springer International Publishing",
address="Cham",
pages="600--610",
isbn="978-3-030-87193-2"
}
